Eigenmodes of Helmholtz¶
Shared by Antoine Rideau
On this page you will find how to show using Castor the eigenmodes of the wave equation on a rectangle with fixed edges.
We start from the following D’Alembert equation on \(\Omega = \left [ 0, x_{0} \right ] \times \left [ 0, y_{0} \right ]\) with Dirichlet boundary condition on \(\Gamma = \partial \Omega\)
considering
gives the Helmholtz equation
L is discretized with dx steps which results in the meshgrid described by X and Y// Parameters
matrix<> L = {1, 2}; // Dimensions
double dx = 0.05; // Space discretization
// Discretization
matrix<> X, Y;
std::tie(X, Y) = meshgrid(colon(0, dx, L(0)), colon(0, dx, L(1)));
See meshgrid.
Laplacian¶
The Helmholtz equation can now be written in vector form
where K stands for the matrix of the Laplacian operator.
The Laplacian operator can be approximated as
This expression leads to this form for K
// Laplacian
long nx = size(X, 2), ny = size(X, 1);
matrix<> e = ones(nx * ny, 5);
e(row(e), 2) = -4;
matrix<> K = full(spdiags(1. / (dx * dx) * e, {-nx, -1, 0, 1, nx}, nx * ny, nx * ny));
See label-spdiags
i such as X(i)==0, X(i)==L(0), Y(i)==0 and Y(i)==L(1) .// Penalization on boundary (Homogeneous Dirichlet condition)
matrix<std::size_t> Ibnd;
Ibnd = find((X == 0) || (X == L(0)) || (Y == 0) || (Y == L(1)));
K(sub2ind(size(K), Ibnd, Ibnd)) = 1e6;
Analytical solution¶
An eigenmodes is caracterize by 2 positive integers \(m\) and \(n\) . Thus the eigenvalues are
and the corresponding eigenmode are
// Analytical
auto Dth = zeros(nx, ny);
for (int m = 0; m < nx; m++)
{
for (int n = 0; n < ny; n++)
{
Dth(m, n) = M_PI * sqrt(pow((m + 1) / L(0), 2) + pow((n + 1) / L(1), 2));
}
}
See zeros .
Eigenmodes¶
Once the Laplacian matrix have been built , eigenvalues are easily acquired in the 1 by nx*ny vector D and eigenvectors in the nx*ny by nx*ny matrix V using the eig function
// Numerical eigen values and vectors
matrix<std::complex<double>> D, V;
std::tie(D, V) = eig(-K, "right");
See eig .
The eigenvalues considerated are these with an imaginary part null and a real part minimal. To do so eigenvalues and eigenvectors are sorted by ascending eigenvalues.
// Sort
matrix<std::size_t> I;
I = argsort(abs(real(D)));
D = eval(D(I));
V = eval(V(row(V), I));
matrix<std::size_t> Ith;
Ith = argsort(Dth);
Dth = eval(Dth(Ith));
Then for each eigenmodes f , only the real part of the corresponding eigenvector is taken.
// Visu
std::vector<figure> fig(5);
for (int f = 0; f < fig.size(); f++)
{
matrix<double> Z = reshape(real(eval(V(row(V), f))), size(X, 1), size(X, 2));
mesh(fig[f], X, Y, Z);
}
Code¶
Here you have all the code at once :
#include "castor/matrix.hpp"
#include "castor/smatrix.hpp"
#include "castor/linalg.hpp"
#include "castor/graphics.hpp"
using namespace castor;
int main(int argc, char const *argv[])
{
// Parameters
matrix<> L = {1, 2}; // Dimensions
double dx = 0.05; // Space discretization
// Discretization
matrix<> X, Y;
std::tie(X, Y) = meshgrid(colon(0, dx, L(0)), colon(0, dx, L(1)));
// Visu mesh
figure fig1;
mesh(fig1, X, Y, zeros(size(X)));
// Laplacian
long nx = size(X, 2), ny = size(X, 1);
matrix<> e = ones(nx * ny, 5);
e(row(e), 2) = -4;
matrix<> K = full(spdiags(1. / (dx * dx) * e, {-nx, -1, 0, 1, nx}, nx * ny, nx * ny));
// Penalization on boundary (Homogeneous Dirichlet condition)
matrix<std::size_t> Ibnd;
Ibnd = find((X == 0) || (X == L(0)) || (Y == 0) || (Y == L(1)));
K(sub2ind(size(K), Ibnd, Ibnd)) = 1e6;
// Analytical
auto Dth = zeros(nx, ny);
for (int m = 0; m < nx; m++)
{
for (int n = 0; n < ny; n++)
{
Dth(m, n) = M_PI * sqrt(pow((m + 1) / L(0), 2) + pow((n + 1) / L(1), 2));
}
}
// Numerical eigen values and vectors
matrix<std::complex<double>> D, V;
std::tie(D, V) = eig(-K, "right");
// Sort
matrix<std::size_t> I;
I = argsort(abs(real(D)));
D = eval(D(I));
V = eval(V(row(V), I));
matrix<std::size_t> Ith;
Ith = argsort(Dth);
Dth = eval(Dth(Ith));
// Visu
std::vector<figure> fig(5);
for (int f = 0; f < fig.size(); f++)
{
matrix<double> Z = reshape(real(eval(V(row(V), f))), size(X, 1), size(X, 2));
mesh(fig[f], X, Y, Z);
}
// Results
std::cout << "-- Numerical eigenvalues --" << endl;
disp(sqrt(real(eval(D(range(0, fig.size()))))), 1, fig.size());
std::cout << "-- Analytical eigenvalues --" << endl;
disp(eval(Dth(range(0, fig.size()))), 1, fig.size());
std::cout << "-- Relative errors --" << endl;
auto errRelative = abs((sqrt(real(eval(D(range(0, fig.size()))))) - eval(Dth(range(0, fig.size())))) / eval(Dth(range(0, fig.size())))) * 100;
disp(errRelative, 1, fig.size());
drawnow(fig1);
return 0;
}
With this code you should get these outputs :
-- Numerical eigenvalues --
Matrix 1x5 of type 'd' (40 B):
3.50946 4.43845 5.65288 6.45167 7.00046
-- Analytical eigenvalues --
Matrix 1x5 of type 'd' (40 B):
3.51241 4.44288 5.66359 6.47656 7.02481
-- Relative errors --
Matrix 1x5 of type 'd' (40 B):
0.08379 0.09980 0.18906 0.38427 0.34671
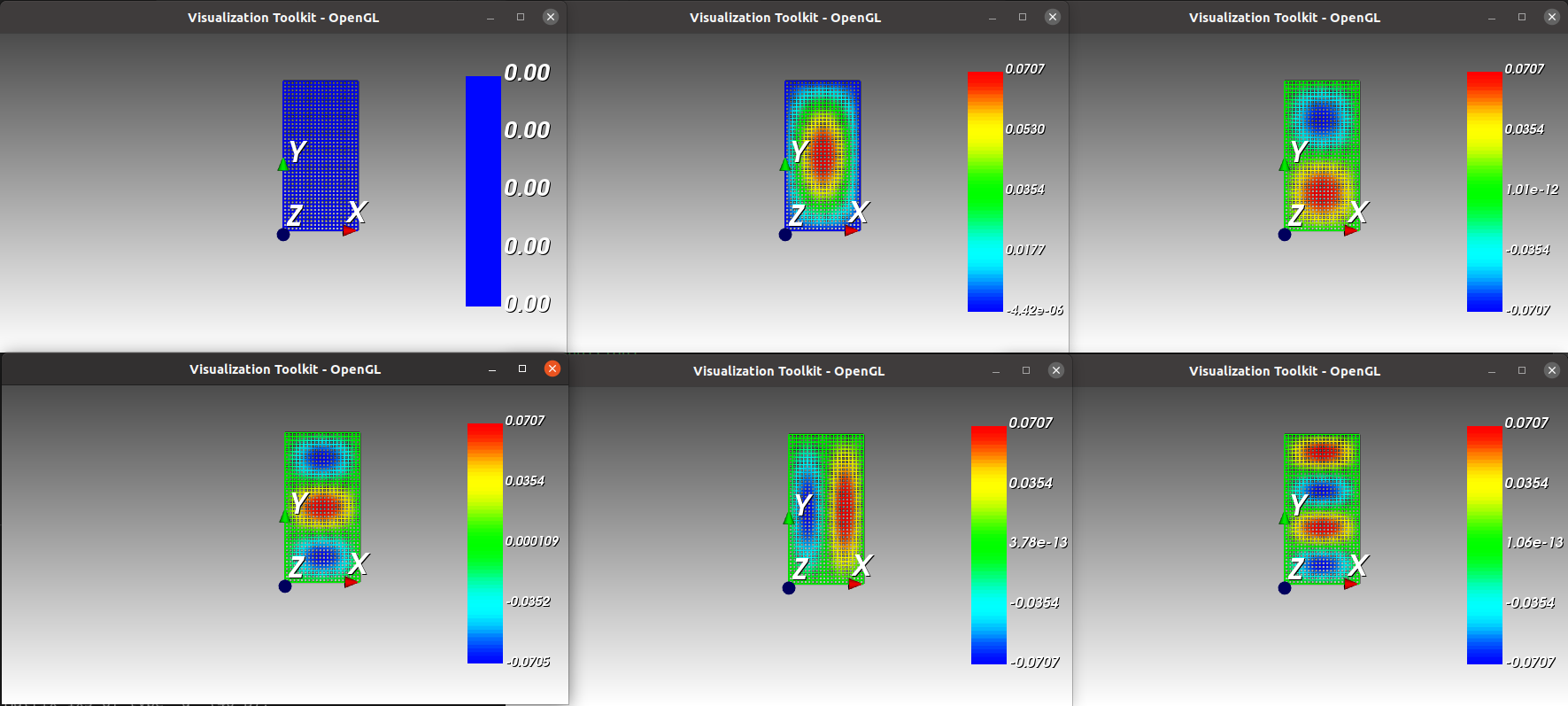
From up left corner to bottom right corner : the meshgrid and the five first eigenmodes.¶
References¶
http://ramanujan.math.trinity.edu/rdaileda/teach/s14/m3357/lectures/lecture_3_4_slides.pdf